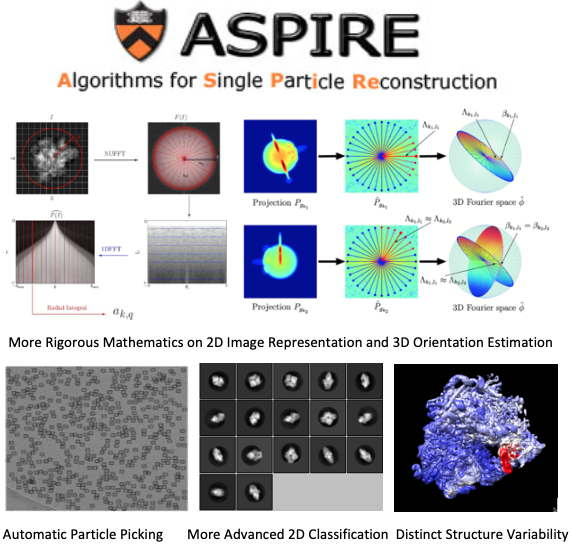
Background on ASPIRE (Algorithms for Single Particle Reconstruction)
Significant progress on computational algorithms and software is one of the major reasons leading the revolution of resolution in three dimensional structure determination of biomolecules using CryoEM, a technique projecting rapidly frozen and randomly orientated 3D particles into 2D noisy images on micrographs and reconstructing 3D density maps in atomistic resolution through computer software. Due to many crucial roles of 3D biomolecules such as protein enzymes for further study in structural and chemical biology, biophysics, biomedical and other related fields, the 2017 Nobel prize in chemistry was awarded to three scholars for significantly advancing the CryoEM technique as explained in this Youtube video.
During the past 10 years, Professor Amit Singer’s group has proposed many new ideas in various numerical algorithms and developed the ASPIRE Matlab package to tackle many problems involved in reconstructing a 3D CryoEM map of biomolecule from corresponding 2D particle images, including CTF estimation, denoising, particle picking, 2D and 3D classification, and ab initio 3D reconstruction.