Background on APPLE-Picker
APPLE-Picker is a submodule of ASPIRE Python package in development for reconstructing a 3D CryoEM map of biomolecule from corresponding 2D particle images, developed by the researchers in Professor Amit Singer’s group. It is an automatic tool to select millions of particles from thousands of micrographs, a critical step in the pipeline of CryoEM image reconstruction. It used to be performed manually but can be very tedious and difficult especially for small particles with low contrast (low signal-noise ratio). The CPU version takes ~80 seconds on average to finish processing one micrograph. To achieve the goal of finishing thousands of micrographs in a few minutes, we need an alternative method, such as GPU accelerating.
2019 Princeton GPU Hackathon
Princeton university held its first GPU hackathon on campus this summer from June 24 to 28, organized and hosted by the Princeton Institute for Computational Science and Engineering (PICSciE), and co-sponsored by NVIDIA and the Oak Ridge Leadership Computing Facility (OLCF). The main goal of this Hackathon was to port research codes to GPUs or optimize them with the help of experts from industry, academia and national labs, as emphasized by Ian Cosden, one of lead organizers and manager of Princeton’s Research Software Engineering Group. This blog reports our attempts and the story behind accelerating APPLE-Picker using GPU and parallel computing in Python.
Computational CryoEM team
When I heard this event at first, I was wondering how we can turn a CPU program into a GPU version just in a few days. My previous field of biomolecular simulation has several leading software packages enabled by GPU computing such as AMBER and GROMACS but most of them ended up implementing complicated GPU kernels of their own. For a previous project I also spent several months on porting high-level GPU library such as openMM into an in-house simulation package mixed with Fortran and C. Accomplishing in a few days looked like mission impossible. However, the ASPIRE package I primarily work on does need GPU support in the future although current redevelopment has not come to this stage yet. At the same time several research software engineers in the Department of Research Computing, including me, also wanted to learn how GPU codes can be implemented in Python. So we decided to give it a try and organized the Computational CryoEM team to join the GPU Hackathon event.
The whole Python package of ASPIRE is still under development but the APPLE-Picker submodule for particle picking is quite ready as a CPU version. It takes the very first position in the whole pipeline of 3D CryoEM map reconstruction but is relatively independent to other modules. Due to those reasons this submodule was an ideal case to work with. The whole Computational CryoEM team included the original developer, Ayelet Heimowitz, and four RSEs, Vineet Bansal, Troy Comi, David Turner, and me.
Roles of RSEs in accomplishing mission
Two weeks before the Hackaton, Ayelet and I had the first meeting to discuss the algorithm and possible hotspots to implement in a GPU version. After that, Vineet and I discussed and completed other initial work such as profiling the APPLE-Picker CPU version, preparing the unit tests, refactoring codes for simpler structure and easier understanding, building necessary PyPI libraries on different computing platforms. After all initial preparations were done, we ended up with a subpackage ready to install and test on major platforms just by several command-line operations.
One week before the Hackaton, all five team members had a one-hour meeting with junior mentor Jonathan Halverson from the RC department and senior mentor Robert (Bob) Crovella from NVIDIA to discuss possible solutions and prepare reading materials, based on our initial profiling of APPLE-Picker. Our initial analysis found that there were three major battlenecks to focus: 1) query_score function including its subfunctions (majorly FFT transform) which took ~83% of the total CPU time; 2) run_svm function and its subfunctions (majorly sklearn functions) which took ~8%; and 3) morphology_ops function and its subfunctions (majorly scipy.ndimage functions) which took ~7%. So all primary bottlenecks were from related external libraries instead of internal implementation. During the meeting we decided to perform an initial search on available GPU implementations such as CuPy, CUFFT, Pyculib, and preparare some general knowledge of Python/Numba and CUDA programming.
On the first morning of the Hackathon, we verified the profiling result, discussed the initial study for possible solutions, decided the initial strategy of replacing Pyfftw lib with CuPy.fft and sklearn SVM lib with ThunderSVM due to the popularity and convenience, and found the possibility of running in multi-GPU mode using multi-process and multi-node. Although everyone participated in the discussion and planning and while there was no absolute distinguishing who should do what part of the whole project, we did split roughly the tasks across the team members to optimize the completion time: Troy took care of cupy.fft; David was responsible for ThunderSVM; Vineet worked on multi-process and multi-node issues and consulted for general coding; Ayelet answered questions on algorithms and the original CPU codes; Jonathon provided support for debugging, profiling, and HPC resources; Bob educated team memberson general knowledge of CuPy and GPU kennels and contacted with remote NVIDIA experts; I organized meetings to discuss possible issues, alternative solutions, and new goals to reach, as well as collecting test results, conducting performance analysis, and preparing daily updates for the Hackathon.
The major progress for each day can be summarized shortly as below: 1) On day one we replaced all Pyfftw codes with CuPy.fft, performed an initial attempt of ThunderSVM, and accelerated the query_score function by a factor of ~4.4 and the whole program by ~2.7. 2) On day two we switched many Numpy functions in query_score and its subfunctions to their corresponding CuPy functions, accelerated the function by a factor of ~22, reached the improvement factor limit of ~5 for the whole program, and the solution for two other bottlenecks further accelerating things by a factor of greater than 10. 3) On day three we replaced the morphology_ops function from the scipy package with a customized GPU kernel (an improvement factor of ~14), solved the reproducible problem of substituting the sklearn SVM with ThunderSVM (an improvement factor of ~3), and accelerated the whole program by a factor of ~11 (~18 for a more aggressive SVM algorithm). 4) On day four we explored the multiprocess functionality by combining the different numbers of GPUs with CPUs for processing one micrograph to optimize the usage of computing resources within a node, enabled the ability of high throughput computing for a large number of micrographs using snakemake, although MPI mode was also discussed. 5) On the morning of day five we cleaned up the codes and wrapped up results for the final report. The figure included below shows in more detail the development iterations during the GPU Hackathon.
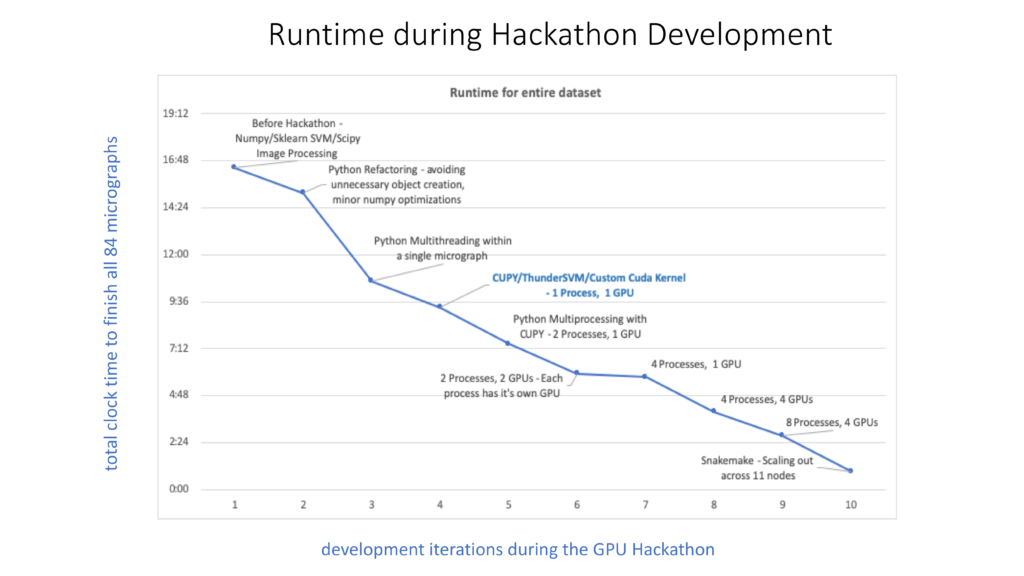
Within a five-day Hackathon event, the RSEs not only explored the various scenarios to accelerate a pure CPU-based Python program from GPU to parallel and concurrent programming in Python, but also led the major roles in refactoring existing codes, developing new GPU codes, and optimizing and performing analysis based on their expertise. “This is could be several months or years work for a researcher only with junior programming experience. The achievement is much more than I expected.” said Ayelet. From my point of view, the successful story has several crucial contributions: 1) We did a very good initial evaluation of the code and performed corresponding preparations comprehensively in order to dive into the phase of solving problem directly at the beginning of Hackathon. 2) We built an excellent team with diverse expertise in mathematical algorithm, Python coding, high performance computing, and GPU programming, to reduce the learning curve and shorten the development cycle significantly in a well-organized and cooperative way. I greatly appreciate this opportunity and well support from Princeton University, OLCF and NVIDIA. I also believe that such an example of solving a coding problem in a few days can be another category of useful RSE activities in addition to general help sessions (which last hours) and projects led by single RSE (which last a few months or longer).
