Background on ASPIRE (Algorithms for Single Particle Reconstruction)
Significant progress on computational algorithms and software is one of the major reasons leading the revolution of resolution in three dimensional structure determination of biomolecules using CryoEM, a technique projecting rapidly frozen and randomly orientated 3D particles into 2D noisy images on micrographs and reconstructing 3D density maps in atomistic resolution through computer software. Due to many crucial roles of 3D biomolecules such as protein enzymes for further study in structural and chemical biology, biophysics, biomedical and other related fields, the 2017 Nobel prize in chemistry was awarded to three scholars for significantly advancing the CryoEM technique as explained in this Youtube video.
During the past 10 years, Professor Amit Singer’s group has proposed many new ideas in various numerical algorithms and developed the ASPIRE Matlab package to tackle many problems involved in reconstructing a 3D CryoEM map of biomolecule from corresponding 2D particle images, including CTF estimation, denoising, particle picking, 2D and 3D classification, and ab initio 3D reconstruction.
Urgent need of redevelopment
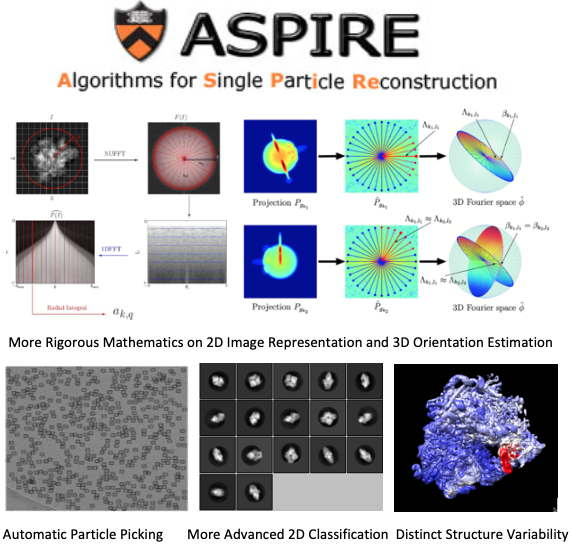
Currently the Matlab ASPIRE package consists of more than 100,000 lines of code, developed by dozens of graduates and postdoc researchers for a single purpose: prototyping their novel ideas and obtaining publishable results for theses and research papers (more than 40 since 2008). The Matlab package includes many algorithm advances in different aspects of CryoEM 3D map reconstruction such as fully automatic particle picking, rigorous mathematics and algorithms for 2D image representation and 3D orientation estimation, accurate 2D classification for images and 3D classification for density maps, and distinct structure variability analysis.
Due to the short-term nature of individual researchers, the Matlab package was developed in a very free style. There does not exist a systematic well-organized framework for laying out the whole package (a very common character for academic software since in many cases a research project includes many trails and failures and can end in an unexpected way). In addition, many good practices in scientific software development were not well followed: Non-meaningful variable names leads to difficulty for understanding; Reusing codes by copying from different developers generates a redundancy problem and makes debugging, optimizing, and maintaining difficult; Lack of usage examples and guides prevents the whole package from being widely distributed to external users; No developer guides and unit tests discourages further developments from within and outside group.
The whole package has been released several times and while there are hundreds of downloads by external groups since the initial release in 2014, there are few usage reports, indicating wide external interests but lack of successful applications. The current state of Matlab codes also created a reusability problem even within the research group for extending previous projects. New developers tends to write the old functions or modules instead of reusing the existing ones. A reusable and sustainable package is an urgent need.
Roles of research software engineers
Two research software engineers Vineet Bansal and I, Junchao Xia, in the Department of Research Computing have taken the major responsibility in developing a new Python package. Vineet is supported by the Department of Research Computing of OIT and the Center for Statistics Machine Learning. He started the initial code conversion in September 2018 and focuses more on software level developments. I am fully supported by the Program of Applied and Computational Mathematics and joined the project at the beginning of 2019. I communicate with researchers in the group and focus more on mathematics and algorithm level for reproducible Python code from the Matlab counterparts. Under the supervision of Professor Amit Singer and RSE manager Ian Cosden, both RSEs also highly collaborate with Joakim Anden at Flatiron Institute, a former researcher and developer.
Generally RSEs in the RC department perform routine tasks such as solving programming issues upon requests, optimizing existing codes, and consulting researchers to follow best practices in developing scientific software. In addition to those tasks, we are also leading other key roles in developing whole ASPIRE Python package, listed as follows but not limited to: 1) Redesign the ASPIRE architecture using the top to down method to fit the general workflow of reconstructing CryoEM 3D. 2) Reorganize the exist code and redevelop new code by following object-oriented programing and design patterns. This involves literature survey for previous work, discussion with researchers once per week for a few months, generating new class hierarchical diagrams and pinpointing corresponding codes in the old package. 3) Develop unittests, usage examples, guiding manuals and other documents for mature codes. 4) Create and maintain a new Github repository including automatic building tests on different platforms (Mac, Windows, and Linux), reporting coverage, and generating online documents. 5) Unify the Python coding style and docstring for current and future developers. 6) Train researchers in developing code using open source software style through git and Github. The new package is under development and can be found in the new link.
Some thoughts from RSE
Although I received PhD in physics, my previous work was highly related to computing, developing computational algorithms and academic software packages for molecular dynamics simulations of biomolecules using high performance computing clusters and heterogeneous distributed volunteer grids. The academic nature of research work, however, made me closer to a computational scientist primarily focusing on algorithm level instead of a software engineer targeting more robust and sustainable software. I appreciate very much the opportunity in this project to sharpen my skills in software engineering such as git for source control, Github for git repository and other automatic tools for improving coding productivity, Pycharm and corresponding development tools for Python, and many others related to the whole life-cycle of professional software development. A great asset working at Princeton is that the Department of Research Computing has a specific series of training events for continuing education and experienced expert team for consulting different problems of HPC computing and software development.
As a research software engineer, I also feel that the “research” component is of importance at my side and there is a unique feature compared to a regular software engineer. In addition to pure coding for software development, my work also requires me to understand algorithms in order to re-implement or debug existing codes. Frequent communication with current researchers is one way. Literature survey is the other inevitable one when the direct consulting is not available. During the redevelopment of a research package that accumulated from many years of effort from previous researchers, the scenario is not unusual. I am happy that my background in physics and computational algorithms is a great help to go through related mathematical equations and algorithms.
